OUR CORE CAPABILITIES

The University of Texas at Austin is home to an almost unique capabi
lity: the ability to quickly and readily synthesize genes by custom order. The Gene Synthesis Facility at the Applied Research Laboratories (ARL) can synthesize long (1-5 kb) DNA fragments of arbitrary sequence from synthetic oligonucleotides in a high-throughput manner. We are currently set up to design, assemble, and test approximately 100 to 200 1-1.5 kb protein-encoding sequences a week, and are working towards routinely building genes and phage sequences up to 5 kb in length.
The generation of novel DNA sequences is frequently a stumbling block in research, as most variant sequences are still produced by PCR cloning and/or mutagenesis of an existing sequence. Such procedures are relatively slow and limited when particular synthetic constructs (fusions, expression) are desired, rather than just the gene itself. Moreover, it is otherwise extremely difficult or expensive to synthesize genes that are codon-optimized or that contain an arbitrarily large set of mutations.
Gene design software developed at UT-ARL can be used to generate gene sequences that are highly optimized for expression from input protein sequences. We begin with a simulated annealing algorithm, restricted by desired protein sequence, which simultaneously optimizes codon usage and AT/GC balance. This routine also includes adjustable fitness parameters for structure, repeated sequences, and arbitrary (user-specified) sequences, allowing for the generation of output DNA sequences maximally optimized for any given application. Our core fabrication software then determines the oligonucleotides necessary to assemble the desired gene sequences and generates the command scripts for robotically controlled assembly using a modified overlap assembly PCR protocol. The general synthetic scheme is that a desired sequence is broken into a set of two to five fragments, each with lengths from 200 to 500 nucleotides. These fragments are then further defined as sets of oligonucleotides, each having a length between 60 and 100 nucleotides. The required oligonucleotides are synthesized in-house with a MerMade 192 DNA synthesizer using chemistry optimized for clean synthesis of oligomers up to 120 nucleotides in length. The synthetic oligonucleotides are then assembled on a Tecan Evo 200 liquid-handling robotic workstation operating from customized scripts generated by the in-house design software to both ensure fidelity and increase throughput. The gene fragments are generated from oligonucleotides using inside-out nucleation reactions. These fragments are subsequently diluted and combined for assembly of the final sequence in a conventional stitch-overlap extension reaction. With the current configuration of the robot, the throughput of this technique is approximately 48 kb of synthesis (48 1-kb genes or 32 1.5-kb genes, for example) per robotic assembly run (4 hours).
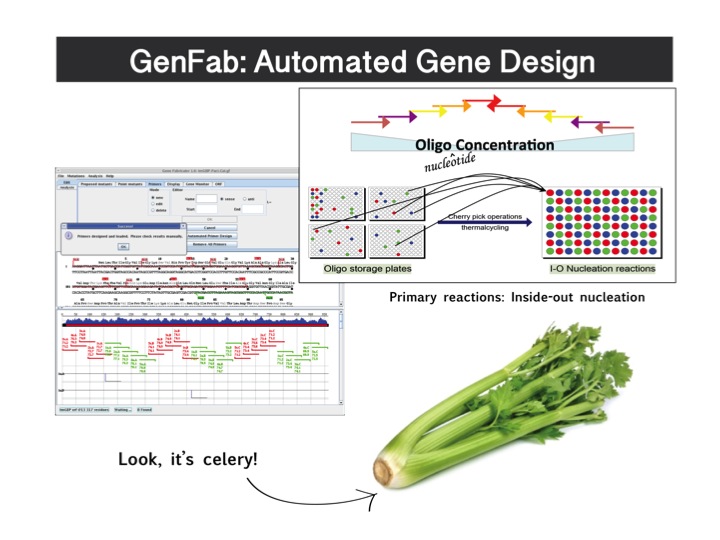
One of the primary hurdles that has been overcome, and that is unique relative to commercial facilities, is the reduction of error rates during synthesis. Oligonucleotide synthesis from phosphoramidites, while robust, yields secondary products primarily due to less than 100% capping efficiency during the polymerization cycle (the N-1 product). These oligonucleotide synthesis side products typically manifest themselves in the assembled gene fragments as single base pair deletions which can result in reading frame-shifting mutations if found within a protein encoding sequence. Additional errors can occur due to DNA polymerase-associated errors (transitions / transversions) during the PCR assembly steps, though these types of errors are less frequent than the single base pair deletions. The reduction of these types of sequence errors while maintaining throughput and cost-effectiveness is a challenge to all projects originating from synthetically derived DNA. We routinely use a mismatch specific endonuclease to cleave (read: eliminate) error-containing sequences and thus enrich for error-free or error-reduced DNA sequences (derived, amusingly, from celery). Using this method we have typically been able to reduce the errors in the synthesized DNA fragments by >6 fold following a single round of treatment.
OUR SERVICE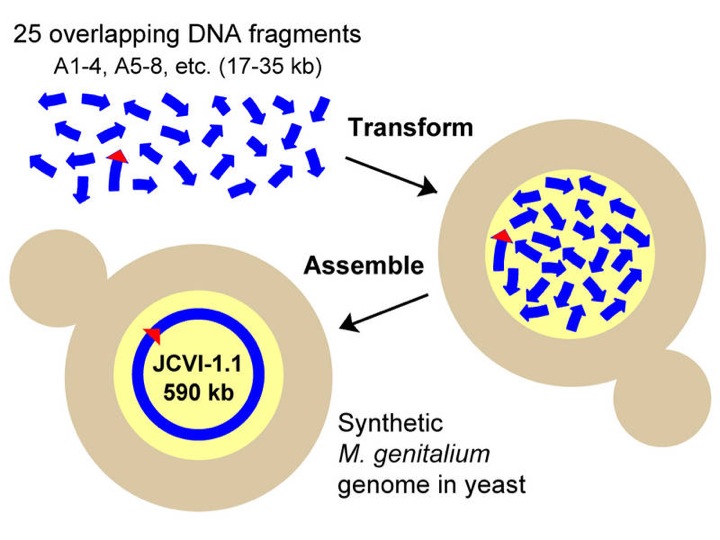
While we typically deliver synthetic pieces uncloned and unsequenced (thus further saving costs to users), correct pieces on the order of 1 kb can typically be found by sequencing only three or so clones. Our pricing for gene synthesis is competitive with industry (yet has all the unique advantages described above) at only $0.22/base.
More importantly, though, we work closely with customers on the design of constructs and can provide useful input regarding how to make fusion constructs, expression constructs, plasmid backbones, codon-optimized genes, codon-optimized libraries, and much more. Please contact Randy Hughes for further details (hughes@mail.utexas.edu).
THINK BIG
As we continue to scale, we are working with CSSB Fellow Dr. Chris Yellman on the yeast-based assembly of even larger constructs by recombination. Yeast can be readily transformed with multiple genes and oligonucleotides and can then efficiently recombine programmed overlaps into larger constructs, including entire plasmids, operons, or even chromosomes (as evidenced by the remarkable construction of the entire genome of Mycobacteria by the Venter Institute). For example, the Marcotte lab is building an entirely humanized proteasome in yeast and we are working with Jon Shimomura-Pierce on the synthesis of artificial C. elegans chromosomes that will allow the animal to mimic Down Syndrome. Contact Dr. Yellman for details (christopher.yellman@gmail.com) and to consult with the Facility about your project.

